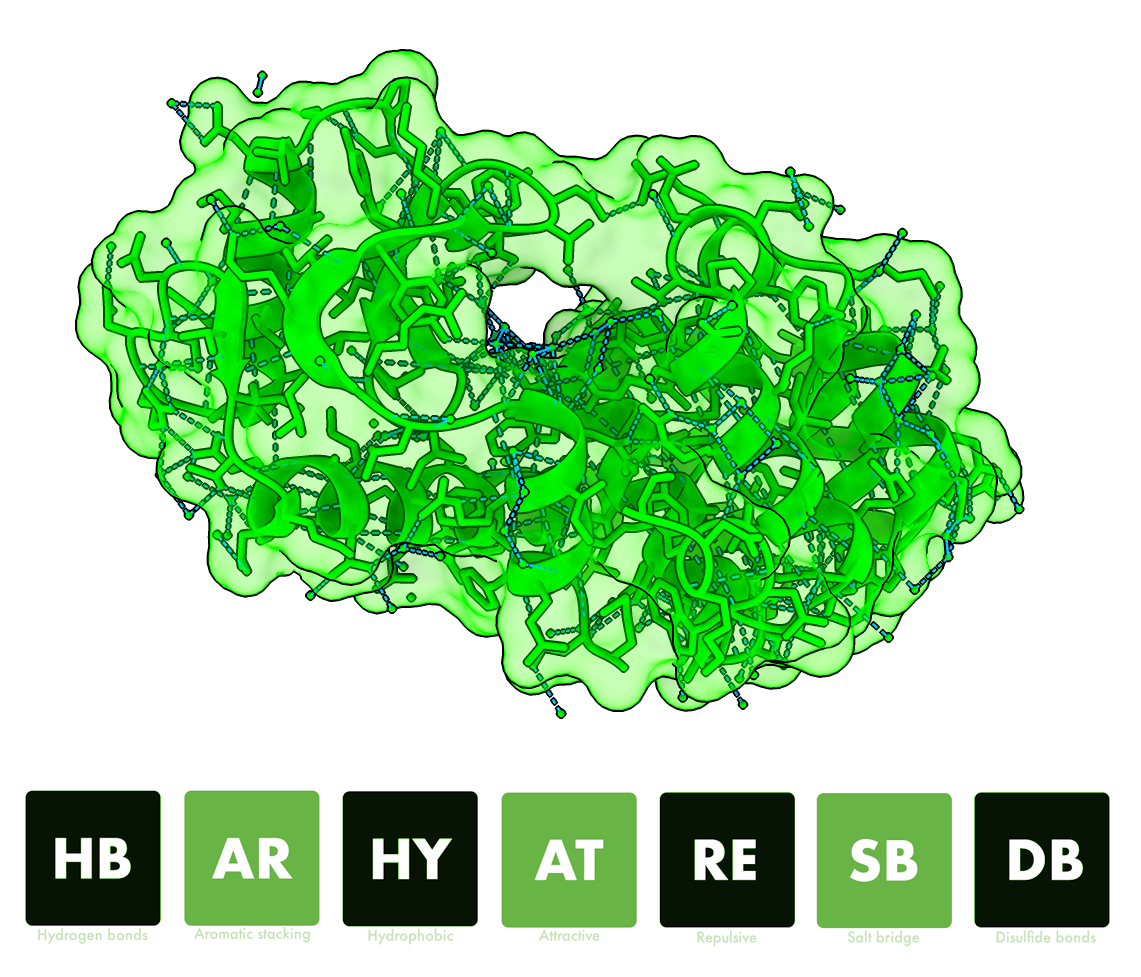
The Interatomic Contact Database & Tool
COCaDA-web is a tool and a database that presents contacts in all structures available in the PDB database. COCaDA calculates seven types of contacts: hydrogen bonds, hydrophobic, aromatic, attractive, repulsive, salt bridges and disulfide bridges.
700,960,631
CONTACTS
649,736,592
INTRA-CHAIN CONTACTS
51,224,039
INTER-CHAIN CONTACTS
233,346
3D STRUCTURES
*Last updated on: ago 17, 2025
How to cite:
LEMOS, Rafael P.; MARIANO, Diego; SILVEIRA, Sabrina A.; MELO-MINARDI, Raquel C. de. COCαDA - Large-Scale Protein Interatomic Contact Cutoff Optimization by Cα Distance Matrices. In: Proceedings of the XVII Brazilian Symposium on Bioinformatics (BSB), 17, p. 59-70, 2024. DOI: https://doi.org/10.5753/bsb.2024.245545
Run COCαDA-web
There are two ways to run COCaDA-web: (1) Upload a file in PDB or CIF format; or (2) enter the 4-digit corresponding PDB code to access COCaDA-db pre-calculated structure contacts. COCaDA-db only supports PDB structures with up to 10,000 amino acid residues. To access structures with more than 10,000 residues, please upload your file.
Cutoffs
Change contact cutoff values (check default values here):
Filter chains
Select the chains you want to calculate contacts for.
AllOnly interchain contacts
Only contacts in the chains:
pH
Change the pH value (default is 7.4):
7.4
Use the pH value defined in the CIF/PDB file

